Estonian Yeast Stock Collection
A collection of yeasts isolated from Estonian nature
The collection was initiated in 2018 when the research project "101 yeast strains from Estonian nature" took place. This project gave an impulse to the study of the distribution of yeast fungi in Estonia. The project was carried out in collaboration with research teams led by Prof. Arnold Kristjuhan and Prof. Tiina Tamm.
The collection has continuously grown over the years. We have been assisted by postgraduate students in Molecular Biosciences who collect samples and isolate new yeast species every year as part of the course "Yeasts - from model organisms to pathogens.
more than 3350 yeast strains isolated from 784 samples;
167 yeast species (of which 144 have been identified at species level, 23 have been identified at family level).
From 2023, the collection belongs to the Global Union of Cultural Collections database (World Data Centre for Microorganisms).
51 students from Estonian schools took part in the collection of natural yeast samples. During the active sampling period from September to October 2018, 822 different strains were isolated. An article was written about the project in the Estonian science news portal "Novaator".
University of Tartu Institute of Molecular and Cell Biology is very thankful to all the students and teachers, who took part in the sample collection!
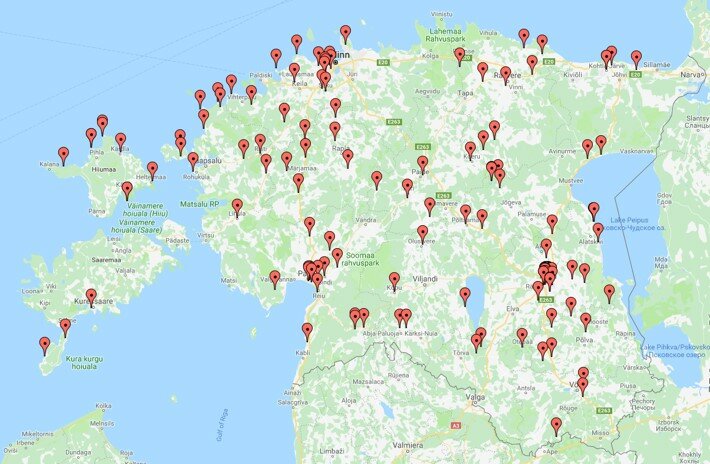
Samples collected during the project on the map of Estonia. Photo: private collection
The following schools participated in the sample collection:
| Audentese Spordigümnaasium | Kääpa Kool | Sindi Gümnaasium |
| Häädemeeste Keskkool | Lihula Gümnaasium | Tallinna Lilleküla Gümnaasium |
| Jõgevamaa Gümnaasium | Nõva Kool | Tallinna Prantsuse Lütseum |
| Kadrina Keskkool | Paide Gümnaasium | Tallinna Reaalkool |
| Koeru Keskkool | Parksepa Keskkool | Tartu Kristjan Jaak Petersoni Gümnaasium |
| Kohila Gümnaasium | Pernova Hariduskeskus | Tartu Tamme Gümnaasium |
| Kohtla-Järve Täiskasvanute Gümnaasium | Põltsamaa Ühisgümnaasium | Tartu Täiskasvanute Gümnaasium |
| Kohtla-Järve Vene Gümnaasium | Pärnu Sütevaka Humanitaargümnaasium | Vinni-Pajusti Gümnaasium |
| Kunda Ühisgümnaasium | Pärnu Ühisgümnaasium | Vändra Gümnaasium |
| Rakvere Gümnaasium |
How do we identify different yeast species?
We use molecular methods to identify the yeast species. For that, genomic DNA is isolated and specific regions from the genome are amplified and sequenced. The resulting sequences are then compared with those in databases. A new species is identified if the region under study differs from previously identified sequences.

On the picture: yeast identification process through molecular methods. Photo taken from personal collection.
The aim of the study was to characterise the culturable epiphytic yeasts associated with apple fruits. Samples were collected in the autumn of 2018 and in the autumn of 2020-2022.
- To isolate the yeasts, apples were collected from 37 different locations in Estonia. The three most frequently analysed local apple varieties were 'Liivi Kuldrenett', 'Antonovka' and 'Talvenauding'.
- During this study, 230 yeast strains were isolated from the samples. These isolates are stored as pure cultures in the Estonian Yeast Stock Collection.
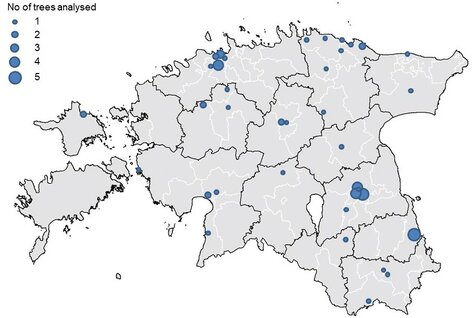
Map of Estonia with locations of sampling sites. The number of trees analysed from each site is shown. Source: Heliyon
Three different regions of the genome were analysed to determine the species identity of the isolates: the 5.8S-ITS region of the ribosomal RNA gene, the D1/D2 region of the ribosomal large subunit RNA and, in some yeasts, the sequence of the translation elongation factor 1α (TEF1) gene. The isolates analysed belonged to 33 different species.
In the seven species isolated, the isolates were significantly divergent from their closest relatives and may therefore represent new distinct species. The study demonstrates that the natural variability of yeasts associated with apple fruits in the northern part of the temperate climate is diverse.
Read more about the study in an article published in Heliyon.
Deoxyribonucleic acid, or DNA, is found in every living cell. Today, DNA analysis is used in many areas of life. To do this, DNA has to be purified from cells and the DNA sequences of interest amplified by polymerase chain reaction (PCR).
Several different methods are used to isolate DNA, some of which are complex and require specialised laboratory equipment. We have developed a method to isolate genomic DNA from different yeast species in a fast (can be done in 1 hour), simple and cost-effective way. The purified DNA can then be used in PCR-based applications.

Steps of the method: growing of yeast cells, extraction of genomic DNA, amplification of specific sequences and analysis of results. Source: STAR Protocols
This method is suitable when a large number of samples need to be analysed simultaneously. The only laboratory equipment required is a heating block and a standard benchtop centrifuge. The whole procedure is carried out in a single tube, no enzymes or harmful chemicals are used, and the necessary solutions can be stored at room temperature.
We used this method for the molecular identification of yeasts isolated from different natural habitats. This method is also suitable for genotyping yeasts used in the laboratory.
For further information, see an article published in STAR Protocols.



