Microbial ecology research group
Nature contains chemical compounds that are degraded by the catabolic activities of microorganisms to generate the energy and to support the world-wide life cycle. Some of the compounds (aromatic and aliphatic hydrocarbons, heavy metals etc.) are highly toxic and mutagenic for organisms. However, several bacteria are able to degrade these compounds. By using natural selection and the molecular genetic engineering methods it is possible to generate supermicrobes applicable for cleaning up of polluted areas.
- Structure, functioning and redundancy of biodegradative pathways and plasmids, evolution of new catabolic features in environmental bacteria
Bacterial catabolic genes are located on extrachromosomal plasmids and/or in chromosomes. Our special focus is on the structure and functioning of these genes and operons. We perform nucleotide sequencing of whole genomes of bacteria in order to understand how these catabolic structures have evolved. We are also interested in the functional redundancy of catabolic modules to verify the best catabolic activities both in indigenous and laboratory constructed bacterial strains. Essential part of this work is the determination of enzyme activities and identification of intermediates of the catabolic pathways and studying the role of these intermediates in the performance of catabolic functions.
- Microbial ecology of the Baltic Sea and adjoining polluted oil shale industry areas
The Baltic Sea is unique among the seas of the world, characterized with the busiest maritime traffic, a high population along its coast area, and the specific unique ecosystem. Large rivers from highly industrialized and agriculturally intensive countries bring high loads of agricultural nutrients (like nitrogen and phosphorous) and hazardous compounds (herbicides, antibacterial agents, oil products, phenolic chemicals etc.) into the Baltic Sea. Therefore our latest research topic has been the elucidation of the biodegradative potential of microbial communities in the Baltic Sea sediment and surface water samples. Culture dependent and independent methods have been used to provide a more accurate picture about the complex microbial communities. Some of the isolated bacterial strains have extremely interesting characteristics useful for the practical purposes in degradation of crude oil derived petroleum hydrocarbons. That is why in these cases we are generating special research projects based on particular strains. This work is done in collaboration with research institutions from Finland and Russia. Another project is ongoing with researchers from Portugal and India to analyze the natural attenuation capacity of the wastewater treatment plant of crude oil refinery and find ways for improving the degradation process. This project targets a global priority issue, i.e., reuse of industrial wastewater.
- Bioaugmentation performances for cleaning up polluted water and sediments
We have long time successful experiences at bioaugmentation of polluted water and sediments. In case of introducing biomass of laboratory selected bacteria to the open environment the key molecular markers (DNA fingerprints) are determined in sense to follow what happens with those bacteria in open nature. Before the field bioaugmentation experiments the behavior of bacteria in laboratory in microcosm experiments mimicking real pollution conditions is always studied.
- Collection of the environmental and laboratory microbial strains (CELMS)
We are responsible for the Estonian National collection of non-medical environmental and laboratory microbial strains (CELMS, http://eemb.ut.ee/ ). Collection contains great variety of indigenous environmental bacteria characterized by DNA sequences of important catabolic regions, species determining gene sequences. Part of the strains has been subjected to whole genome sequence determination. This is a basic facility for our laboratory research. The new bacterial strains obtained within research projects conducted in our department are stored at the culture collection. The intellectual property agreement between author of the bacterial strain and official representative of collection guarantees author’s rights.
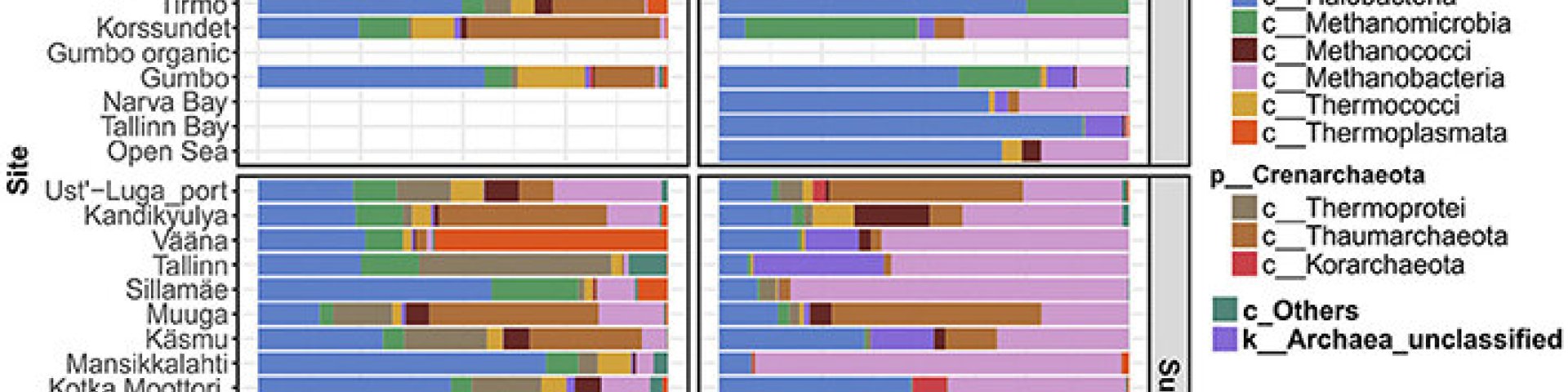
1. Yan, L.; Yu, D.; Hui, N.; Naanuri, E.; Viggor, S.; Gaforov, A.; Sokolov, S.; Heinaru, A.; Romantschuk, M. (2018). Distribution of archaeal communities along the coast of the Gulf of Finland and their response to oil contamination. Frontiers in Microbiology, 9, 1−19.
2. Jõesaar, Merike; Viggor, Signe; Heinaru, Eeva; Naanuri, Eve; Mehike, Maris; Leito, Ivo; Heinaru, Ain (2017). Strategy of Pseudomonas pseudoalcaligenes C70 for effective degradation of phenol and salicylate. PLoS ONE, 12 (3), 1−17.
3. Heinaru, Eeva; Naanuri, Eve; Grünbach, Maarja; Jõesaar, Merike; Heinaru, Ain (2016). Functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene, 589, 90−98.
4. Viggor, Signe; Jõesaar, Merike; Vedler, Eve; Kiiker, Riinu; Pärnpuu, Liis; Heinaru, Ain. (2015). Occurrence of diverse alkane hydroxylase alkB genes in indigenous oil-degrading bacteria of Baltic Sea surface water. Marine Pollution Bulletin, 101 (2), 507−516.
5. Tiirik, Kertu; Nõlvak, Hiie; Oopkaup, Kristjan; Truu, Marika; Preem, Jens-Konrad; Heinaru, Ain; Truu, Jaak (2014). Characterization of the bacterioplankton community and its antibiotic resistance genes in the Baltic Sea. Biotechnology and Applied Biochemistry, 61 (1), 23−32.
(täiendamisel)