-
Faculty of Arts and HumanitiesDean's Office, Faculty of Arts and HumanitiesJakobi 2, r 116-121 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of History and ArchaeologyJakobi 2 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Estonian and General LinguisticsJakobi 2, IV korrus 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Philosophy and SemioticsJakobi 2, III korrus, ruumid 302-337 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Cultural ResearchÜlikooli 16 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of Foreign Languages and CulturesLossi 3 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Theology and Religious StudiesÜlikooli 18 50090 Tartu linn, Tartu linn, Tartumaa EST0Viljandi Culture AcademyPosti 1 71004 Viljandi linn, Viljandimaa EST0Professors emeriti, Faculty of Arts and Humanities0Associate Professors emeriti, Faculty of Arts and Humanities0Faculty of Social SciencesDean's Office, Faculty of Social SciencesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of EducationJakobi 5 51005 Tartu linn, Tartu linn, Tartumaa EST0Johan Skytte Institute of Political StudiesLossi 36, ruum 301 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Economics and Business AdministrationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of PsychologyNäituse 2 50409 Tartu linn, Tartu linn, Tartumaa EST0School of LawNäituse 20 - 324 50409 Tartu linn, Tartu linn, Tartumaa EST0Institute of Social StudiesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Narva CollegeRaekoja plats 2 20307 Narva linn, Ida-Virumaa EST0Pärnu CollegeRingi 35 80012 Pärnu linn, Pärnu linn, Pärnumaa EST0Professors emeriti, Faculty of Social Sciences0Associate Professors emeriti, Faculty of Social Sciences0Faculty of MedicineDean's Office, Faculty of MedicineRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Biomedicine and Translational MedicineBiomeedikum, Ravila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of PharmacyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of DentistryL. Puusepa 1a 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Clinical MedicineL. Puusepa 8 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Family Medicine and Public HealthRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Sport Sciences and PhysiotherapyUjula 4 51008 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Medicine0Associate Professors emeriti, Faculty of Medicine0Faculty of Science and TechnologyDean's Office, Faculty of Science and TechnologyVanemuise 46 - 208 51003 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Computer ScienceNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa ESTInstitute of GenomicsRiia 23b/2 51010 Tartu linn, Tartu linn, Tartumaa ESTEstonian Marine Institute0Institute of PhysicsInstitute of ChemistryRavila 14a 50411 Tartu linn, Tartu linn, Tartumaa EST0Institute of Mathematics and StatisticsNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of Molecular and Cell BiologyRiia 23, 23b - 134 51010 Tartu linn, Tartu linn, Tartumaa ESTTartu ObservatoryObservatooriumi 1 61602 Tõravere alevik, Nõo vald, Tartumaa EST0Institute of TechnologyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Ecology and Earth SciencesJ. Liivi tn 2 50409 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Science and Technology0Associate Professors emeriti, Faculty of Science and Technology0Institute of BioengineeringArea of Academic SecretaryHuman Resources OfficeUppsala 6, Lossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Area of Head of FinanceFinance Office0Area of Director of AdministrationInformation Technology Office0Administrative OfficeÜlikooli 17 (III korrus) 51005 Tartu linn, Tartu linn, Tartumaa EST0Estates Office0Marketing and Communication OfficeÜlikooli 18, ruumid 102, 104, 209, 210 50090 Tartu linn, Tartu linn, Tartumaa EST0Area of RectorRector's Strategy OfficeInternal Audit OfficeArea of Vice Rector for Academic AffairsOffice of Academic Affairs0University of Tartu Youth AcademyUppsala 10 51003 Tartu linn, Tartu linn, Tartumaa EST0Student Union OfficeÜlikooli 18b 51005 Tartu linn, Tartu linn, Tartumaa EST0Centre for Learning and TeachingArea of Vice Rector for ResearchUniversity of Tartu LibraryW. Struve 1 50091 Tartu linn, Tartu linn, Tartumaa EST0Grant OfficeArea of Vice Rector for DevelopmentCentre for Entrepreneurship and InnovationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0University of Tartu Natural History Museum and Botanical GardenVanemuise 46 51003 Tartu linn, Tartu linn, Tartumaa EST0International Cooperation and Protocol Office0University of Tartu MuseumLossi 25 51003 Tartu linn, Tartu linn, Tartumaa EST0
DNA detection of microbial pathogens in human blood
finding rare and poorly studied variations
determination of the composition and origin of food by means of DNA in food
reliable and rapid analysis of genomic sequences
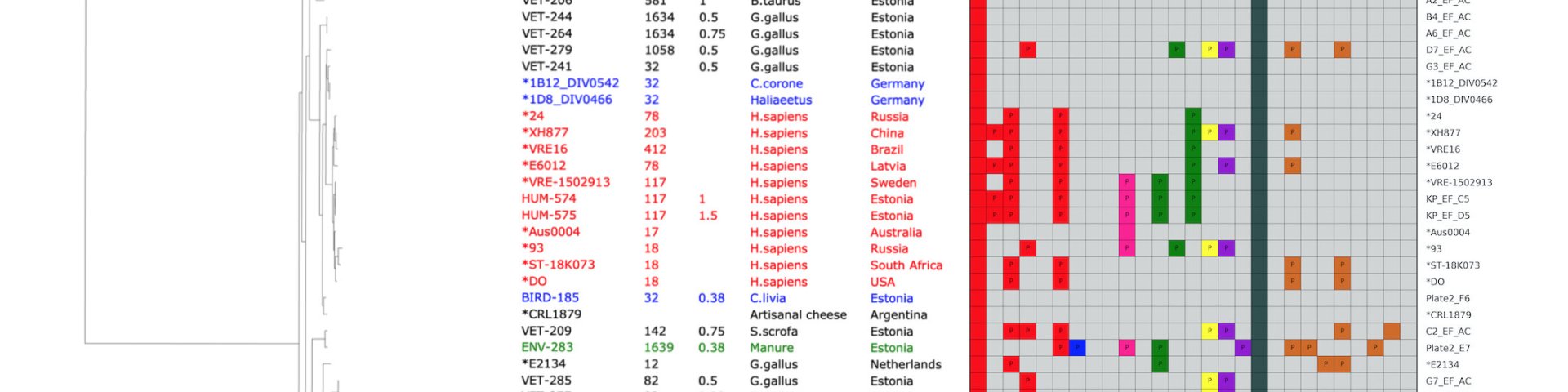
Bioinformatics research group
We develop computational methods for faster and more reliable analysis of genomic sequences. Some examples of such methods are:
* prediction of antibiotic resistance and plasmid content of bacterial isolate from its genomic sequence [1,2];
* detection of microbial pathogens from human cell-free DNA, from wastewater samples and from other relevant metagenomic samples;
* detection of rare variants in personal human genomes [3,4];
* surveillance of food composition and food origin, based on traces of DNA detected from food [5].
Our future plan is to develop a large set of DNA sequence-based predictive models and software for surveillance of bacterial pathogens (virulence, resistance to drugs or cleaning agents), microbiome and food composition.
Oselin K, Kaplinski L, Jääger A, Valter A, Puurand T, et al. (2023). Comparative Genomic Analysis to Distinguish Triple Metachronous Lung Carcinoma from Metastatic Recurrence: Brief Report on First Case Treated with Three Consecutive VATS Lobectomies. Annals of Medical and Clinical Oncology 5 (145)
Pajuste FD, Remm M. (2023). GeneToCN: an alignment-free method for gene copy number estimation directly from next-generation sequencing reads. Scientific Reports, 13 (17765)
Kaplinski L, Möls M, Puurand T, Remm M. (2023). DOCEST—fast and accurate estimator of human NGS sequencing depth and error rate. Bioinformatics Advances, 3 (1)
Kaplinski L, Möls M, Puurand T, Pajuste FD, Remm M. (2021). KATK: Fast genotyping of rare variants directly from unmapped sequencing reads. Human Mutation, 42(6):777-786
Raime K, Krjutškov K and Remm M. (2020). Method for the Identification of Plant DNA in Food Using Alignment-Free Analysis of Sequencing Reads: A Case Study on Lupin. Front. Plant Sci., 11:646
Kaplinski L, Möls M, Puurand T, Pajuste FD, Remm M (2021). KATK: Fast genotyping of rare variants directly from unmapped sequencing reads. Human Mutation, 42(6):777-786
Puurand T, Kukuškina V, Pajuste FD and Remm M. (2019). AluMine: alignment-free method for the discovery of polymorphic Alu element insertions. Mobile DNA, 10:31
Roosaare M, Puustusmaa M, Möls M, Vaher M, and Remm M. (2018). PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ, 6: e4588
Aun E, Brauer A, Kisand V, Tenson T and Remm M. (2018). A k-mer-based method for the identification of phenotype-associated genomic biomarkers and predicting phenotypes of sequenced bacteria. PLoS Computational Biology, 14(10):e1006434


