-
Faculty of Arts and HumanitiesDean's Office, Faculty of Arts and HumanitiesJakobi 2, r 116-121 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of History and ArchaeologyJakobi 2 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Estonian and General LinguisticsJakobi 2, IV korrus 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Philosophy and SemioticsJakobi 2, III korrus, ruumid 302-337 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Cultural ResearchÜlikooli 16 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of Foreign Languages and CulturesLossi 3 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Theology and Religious StudiesÜlikooli 18 50090 Tartu linn, Tartu linn, Tartumaa EST0Viljandi Culture AcademyPosti 1 71004 Viljandi linn, Viljandimaa EST0Professors emeriti, Faculty of Arts and Humanities0Associate Professors emeriti, Faculty of Arts and Humanities0Faculty of Social SciencesDean's Office, Faculty of Social SciencesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of EducationJakobi 5 51005 Tartu linn, Tartu linn, Tartumaa EST0Johan Skytte Institute of Political StudiesLossi 36, ruum 301 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Economics and Business AdministrationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of PsychologyNäituse 2 50409 Tartu linn, Tartu linn, Tartumaa EST0School of LawNäituse 20 - 324 50409 Tartu linn, Tartu linn, Tartumaa EST0Institute of Social StudiesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Narva CollegeRaekoja plats 2 20307 Narva linn, Ida-Virumaa EST0Pärnu CollegeRingi 35 80012 Pärnu linn, Pärnu linn, Pärnumaa EST0Professors emeriti, Faculty of Social Sciences0Associate Professors emeriti, Faculty of Social Sciences0Faculty of MedicineDean's Office, Faculty of MedicineRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Biomedicine and Translational MedicineBiomeedikum, Ravila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of PharmacyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of DentistryL. Puusepa 1a 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Clinical MedicineL. Puusepa 8 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Family Medicine and Public HealthRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Sport Sciences and PhysiotherapyUjula 4 51008 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Medicine0Associate Professors emeriti, Faculty of Medicine0Faculty of Science and TechnologyDean's Office, Faculty of Science and TechnologyVanemuise 46 - 208 51003 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Computer ScienceNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa ESTInstitute of GenomicsRiia 23b/2 51010 Tartu linn, Tartu linn, Tartumaa ESTEstonian Marine Institute0Institute of PhysicsInstitute of ChemistryRavila 14a 50411 Tartu linn, Tartu linn, Tartumaa EST0Institute of Mathematics and StatisticsNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of Molecular and Cell BiologyRiia 23, 23b - 134 51010 Tartu linn, Tartu linn, Tartumaa ESTTartu ObservatoryObservatooriumi 1 61602 Tõravere alevik, Nõo vald, Tartumaa EST0Institute of TechnologyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Ecology and Earth SciencesJ. Liivi tn 2 50409 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Science and Technology0Associate Professors emeriti, Faculty of Science and Technology0Institute of BioengineeringArea of Academic SecretaryHuman Resources OfficeUppsala 6, Lossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Area of Head of FinanceFinance Office0Area of Director of AdministrationInformation Technology Office0Administrative OfficeÜlikooli 17 (III korrus) 51005 Tartu linn, Tartu linn, Tartumaa EST0Estates Office0Marketing and Communication OfficeÜlikooli 18, ruumid 102, 104, 209, 210 50090 Tartu linn, Tartu linn, Tartumaa EST0Area of RectorRector's Strategy OfficeInternal Audit OfficeArea of Vice Rector for Academic AffairsOffice of Academic Affairs0University of Tartu Youth AcademyUppsala 10 51003 Tartu linn, Tartu linn, Tartumaa EST0Student Union OfficeÜlikooli 18b 51005 Tartu linn, Tartu linn, Tartumaa EST0Centre for Learning and TeachingArea of Vice Rector for ResearchUniversity of Tartu LibraryW. Struve 1 50091 Tartu linn, Tartu linn, Tartumaa EST0Grant OfficeArea of Vice Rector for DevelopmentCentre for Entrepreneurship and InnovationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0University of Tartu Natural History Museum and Botanical GardenVanemuise 46 51003 Tartu linn, Tartu linn, Tartumaa EST0International Cooperation and Protocol Office0University of Tartu MuseumLossi 25 51003 Tartu linn, Tartu linn, Tartumaa EST0
• Mitochondrial DNA helicases
Yeast Saccharomyces cerevisiae
Mitochondrial DNA topology and stable inheritance
Mitochondrial DNA helicases
Yeast mitochondrial research group
Mitochondria are organelles that have their own genomic DNA. We are interested in the mechanisms by which the mitochondrial genome is propagated during the cell cycle. We try to understand what changes in the structure of DNA molecules take place during synthesis and how mitochondrial DNA replication and recombination are related. We are also interested in enzymes involved in mitochondrial DNA metabolism - DNA polymerase, which synthesizes mitochondrial DNA; nucleases required for DNA stability and helicases, which are motor enzymes that modulate the structure of nucleic acids.
Analysis of mitochondrial DNA topology and stable inheritance
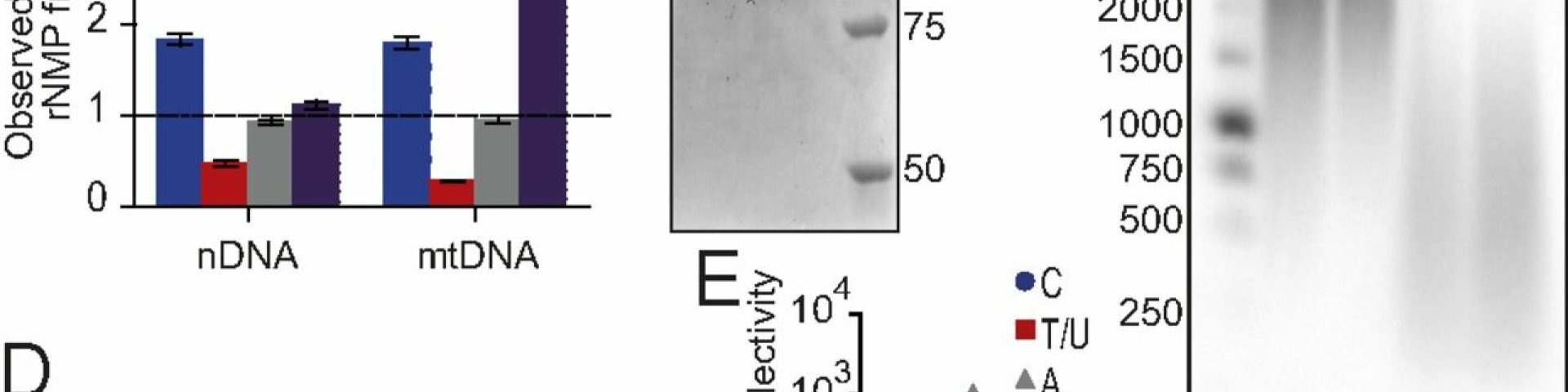
Analysis by our group has shown that mitochondrial DNA forms complex higher-order network structures in yeast and cannot be described by the simple ring structures depicted in the textbooks. Our data indicate that such DNA structures are likely to result from recombination and that yeast mitochondria do not have the specific DNA synthesis initiation structures that should be present when using RNA primers.
The S. cerevisiae model organism allows us to systematically analyze the effects of various enzymes on mitochondrial DNA stability, as baker's yeast can grow without a functional respiratory chain. We have constructed yeast strains in which RNA synthesis in mitochondria is eliminated, allowing the identification of factors important in recombinant DNA synthesis.
Function of mitochondrial DNA helicases
As a rule, the DNA helicase is involved in DNA replication in various systems, and its task is to unravel the strands at the replication fork. As a result of our laboratory work, two DNA helicases have been identified in the mitochondria, but it has also been found that the classical replicative helicase in yeast mitochondria is apparently absent. For functional analysis of mitochondrial DNA helicases, we use a combination of biochemical experiments with purified proteins and in vivo analysis. Thus, we have shown that these mirochondrial helicases have specificity for branched-chain DNA molecules, suggesting their role in recombinant processes.
- Sillamaa, S., Piljukov, V. J., Vaask, I., Sedman, T., Jõers, P., & Sedman, J. (2023). UvrD-like helicase Hmi1 Has an ATP independent role in yeast mitochondrial DNA maintenance. DNA repair, 132, 103582. https://doi.org/10.1016/j.dnarep.2023.103582
-
Tarrés-Solé, A., Battistini, F., Gerhold, J. M., Piétrement, O., Martínez-García, B., Ruiz-López, E., Lyonnais, S., Bernadó, P., Roca, J., Orozco, M., Le Cam, E., Sedman, J., & Solà, M. (2023). Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism. Nucleic acids research, 51(11), 5864–5882. https://doi.org/10.1093/nar/gkad397
-
Piljukov, V. J., Sillamaa, S., Sedman, T., Garber, N., Rätsep, M., Freiberg, A., & Sedman, J. (2023). Mitochondrial Irc3 helicase of the thermotolerant yeast Ogataea polymorpha displays dual DNA- and RNA-stimulated ATPase activity. Mitochondrion, 69, 130–139.
-
Piljukov VJ, Garber N, Sedman T, Sedman J.(2020) Irc3 is a monomeric DNA branch point-binding helicase in mitochondria of the yeast Saccharomyces cerevisiae. FEBS Lett. 594(19):3142-3155.
-
Sedman T, Garber N, Gaidutšik I, Sillamaa S, Paats J, Piljukov VJ, Sedman J. (2017) Mitochondrial helicase Irc3 translocates along double-stranded DNA. FEBS Lett. 591(23):3831-3841.
-
Wanrooij PH, Engqvist MKM, Forslund JME, Navarrete C, Nilsson AK, Sedman J, Wanrooij S, Clausen AR, Chabes (2017) Ribonucleotides incorporated by the yeast mitochondrial DNA polymerase are not repaired. Proc Natl Acad Sci U S A. ;114(47):12466-12471.
-
Gaidutšik I, Sedman T, Sillamaa S, Sedman J (2016) Irc3 is a mitochondrial DNA branch migration enzyme. Sci Rep. 6:26414.
-
Sedman T, Gaidutšik I, Villemson K, Hou Y, Sedman J. (2014) Double-stranded DNA-dependent ATPase Irc3p is directly involved in mitochondrial genome maintenance. Nucleic Acids Res. 42(21):13214-27.


